Mitochondrial protein biogenesis
Mitochondria evolved from the engulfment of a prokaryote by eukaryotic ancestor cell. During the course of evolution, the prokaryotic endosymbiont developed to a cellular organelle that fulfills essential functions for the cell such as energy production, iron-sulfur cluster formation, lipid and heme biosynthesis. The vast majority of the genetic information of the prokaryote was transferred to the host nucleus. Consequently, about 99% of the mitochondrial proteins are synthesized on cytosolic ribosomes as precursors and have to be imported into the organelle in a post-translational manner. The translocase of the outer mitochondrial membrane (TOM complex) forms the entry gate for most of the precursors. Subsequently, specific protein machineries sort the precursors to the different compartments of mitochondria (Figure 1): Outer and inner membrane, matrix and intermembrane space.t.
Biogenesis of the mitochondrial outer membrane
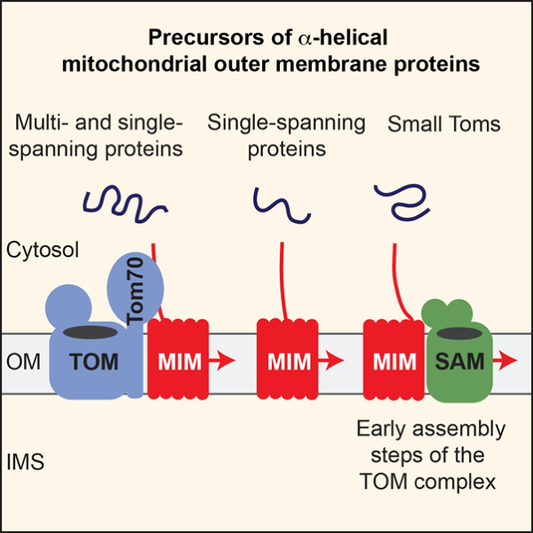
We are particular interested in the biogenesis of mitochondrial outer membrane proteins. These proteins span the membrane by one or more transmembrane a-helices or by a b-barrel structure. b-barrel proteins are imported by the TOM machinery and inserted into the membrane from the intermembrane space side by the sorting and assembly machinery (SAM complex). We could show that the action of both translocases is physically linked to each other to ensure efficient import and folding of the precursor. Small Tim proteins of the intermembrane space shield hydrophobic segments and stabilize the binding of the precursor to the TOM-SAM supercomplex.
We have identified a new import pathway for proteins with a single and multiple transmembrane a-helixes. The import of some of these proteins involves by TOM receptors on the mitochondrial surface, but most of the proteins do not pass the TOM channel. Instead, the mitochondrial import machinery (MIM complex) promotes insertion and assembly of the proteins in the outer mitochondrial membrane (Figure 2). The MIM machinery can also cooperate with the SAM complex in the assembly of the TOM complex. We aimed to characterize the molecular mechanism of this novel protein import pathway.
Molecular network of mitochondrial protein translocases
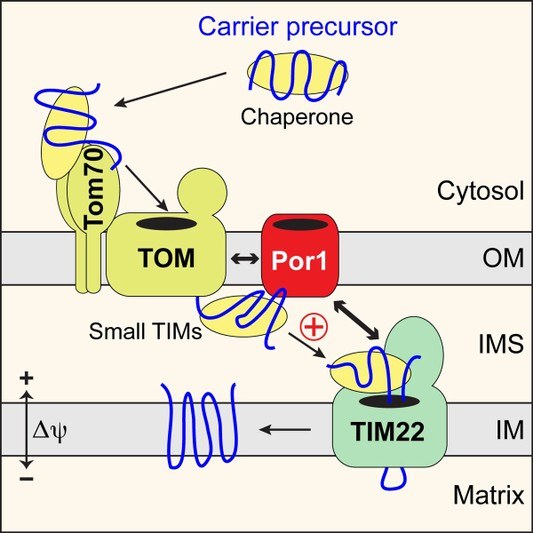
Our study revealed that outer membrane protein translocases associate with many partner proteins, including exciting links to mitochondrial metabolism and membrane contact sites. We found a role of the yeast voltage-dependent anion channel (VDAC), porin, in the import of carrier proteins into the inner membrane. Porin functions as coupling factor that coordinates protein import steps at both membranes. Thus, porin plays a dual role in mitochondrial biogenesis (Figure 3). First, it functions as channel for the flux of metabolites and ion across the outer membrane. Second, porin promotes import of carrier proteins by spatially organizing outer and inner membrane transport steps.
Protein translocases are intimately linked to organelle contact sites. TOM subunits associate with ER-resident proteins. Furthermore, a fraction of the sorting and assembly machinery (SAM complex) binds Mdm10 that is also a central component of the endoplasmic reticulum encounter structure (ERMES) that forms a molecular bridge between the outer membrane and the endoplasmic reticulum (ER). ERMES facilitates lipid transfer between both organelles. We demonstrated how Mdm10 couples with two distinct protein machineries. Thus, both examples illustrates that protein transport is connected to metabolite flux and lipid biogenesis. Our goal is to understand how protein translocases are integrated into a protein network that control mitochondrial biogenesis and function.
Quality control of mitochondrial protein biogenesis
Protein import has to be tightly controlled. Defects in protein import cause massive cellular stress and can eventually lead to cell death. Thus, quality mechanisms have to monitor mitochondrial protein biogenesis at different steps. This includes an intricate network of molecular chaperones that transports precursor proteins to the mitochondrial surface. Hsp70 chaperones are central players in protein targeting. Co-chaperones like J-proteins control protein targeting to mitochondria. We found that distinct J-proteins binds to different TOM receptors, suggesting that protein transport starts in the cytosol.
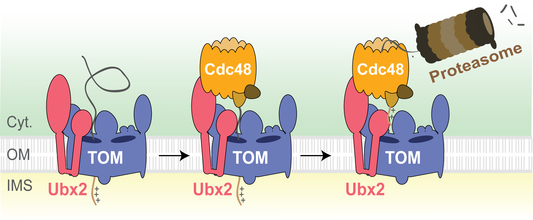
Precursor proteins that prematurely fold can arrest in protein translocases such as the TOM complex during protein import into mitochondria, leading to clogging of protein translocases. We identified a novel mechanism that clears clogged TOM complexes, the protein translocation-associated degradation (mitoTAD) pathway (Figure 4). The protein Ubx2 associates with the TOM complex and forms a docking site for the cytosolic AAA ATPase Cdc48. Cdc48 extracts stalled precursor proteins from the TOM complex and facilitates their proteasomal degradation. Our goal is to understand the molecular mechanisms that govern mitochondrial proteostasis at the mitochondrial outer membrane.


